02 December 2021: Clinical Research
Copy Number Variation Analysis of IL22 and LCE3C in Different Subtypes of Psoriasis in a Chinese Han Population
Caihong ZhuDOI: 10.12659/MSM.934927
Med Sci Monit 2021; 27:e934927
Abstract
BACKGROUND: Psoriasis is a chronic, immune-mediated and hyperproliferative skin disease with both genetic and environmental components. Copy number variations (CNV) of IL22 and LCE3C-LCE3B deletion have been confirmed to be predisposed to psoriasis vulgaris (PsV) in several ethnic groups. However, it remains to be clarified whether CNVs of IL22 and LCE3C are associated with different subtypes of psoriasis (psoriatic arthritis, PsA; erythrodermic psoriasis, EP; and generalized pustular psoriasis, GPP).
MATERIAL AND METHODS: We enrolled 897 Han Chinese individuals, including 478 patients and 419 healthy controls, and detected CNVs of IL22 and LCE3C using the comparative CT method by real-time PCR, and Pearson’s χ² test was used to evaluated the copy number difference among subtypes.
RESULTS: CNVs of IL22 were significantly higher in PsV than in healthy controls (P<0.001). CNV of LCE3C in PsV, PsA, and GPP groups were significantly lower compared to healthy controls. When linked with clinical parameters, mild psoriasis carried less IL22 copy numbers than that in severe psoriasis (P=0.043). Neither IL22 or LCE3C CNVs were associated with age of onset.
CONCLUSIONS: CNVs of LCE3C and IL22 might differentially contribute to subtypes of psoriasis. These findings suggest complex and diverse genetic variations in and among different clinical subtypes of psoriasis.
Keywords: Body Surface Area, DNA Copy Number Variations, Psoriasis, Cornified Envelope Proline-Rich Proteins, Female, Genetic Predisposition to Disease, Humans, Interleukins, Male
Background
Psoriasis is an autoimmune-mediated disease that primarily affects the skin, nails, and joints [1,2]. The disease presents mainly in 4 presentations, and classifications have been proposed based on clinical course. PsV, characterized by erythematous and scaly plaques, is usually found on the elbows, knees, scalp, and back and affects 85–90% of patients with psoriasis [3–6]. PsA is a type of psoriasis that is associated with erosive polyarthritis and usually has a negative serologic test result for rheumatoid factor; it shares some common genetic background with psoriasis [6,7]. EP presents as widespread erythema with thick scales (exfoliation of the skin, which is quite different from the thick, adherent, and white scales of PsV) [5,6,8]. GPP is characterized by infiltration of neutrophil granulocytes in the epidermis to such an extent that clinically visible sterile pustules develop [5,6,9,10]. Both EP and GPP are severe subtypes of psoriasis that affect nearly the entire body surface. Approximately 30% of psoriasis patients develop PsA, EP, or GPP. Even though these clinical phenotypes share some common manifestations, their molecular mechanisms differ and remain to be classified further.
It was widely accepted that aberrant secretion of cytokines and skin barrier dysfunction are the 2 key features in psoriasis pathogenesis. Some cytokines, including
CNVs, defined as deletions or duplications of DNA segments longer than 1 kb, cover approximately 12% of the human genome and encompass more nucleotides than do SNPs [14]. CNVs can affect human phenotypes by directly affecting gene expression levels through duplication or deletion of a specific DNA sequence, or by indirectly altering gene expression, such as affecting the transcriptional regulatory elements of corresponding genes [15–18]. Several studies revealed that CNVs are important genetic factors that predispose to psoriasis [13,19–24]. CNVs of
Material and Methods
SUBJECTS:
We enrolled 897 Chinese Han individuals, including 478 patients (74 PsA, 69 EP, 52 GPP, and 283 PsV) and 419 healthy controls. None of the patients in the study had received systemic or physical therapy within the 2 weeks before a blood sample was drawn. All subjects were recruited from the First Affiliated Hospital of Anhui Medical University, China. Psoriasis patients with joint symptoms were confirmed with a diagnosis of PsA according to the Classification for Psoriatic Arthritis criteria [29]. The remaining patients with no joint symptoms were divided into PsV, EP, and GPP groups according to their clinical manifestations which state in the introduction section. All patients were diagnosed according to their individual clinical manifestation and/or histopathological performance by 2 experienced dermatologists (Table 1). Psoriasis patients were grouped as early- and late-onset psoriasis. Early-onset psoriasis is defined as onset before 40 years of age, while the rest are defined as late-onset psoriasis. Psoriasis severity is categorized as mild, moderate, and severe, which is guided by body surface area (BSA). BSA is preferential measurement in the US and other countries in both routine clinical practice and clinical trials [30,31]. BSA was assessed by a trained physician. The severity of psoriasis was classified as follows: mild: BSA ≤3%, moderate: 3%< BSA ≤10%, and severe: BSA >10%. Healthy controls were confirmed to have no personal or family history of autoimmune diseases such as psoriasis, rheumatoid arthritis, and systemic lupus erythematosus. This study was approved by the Ethics Committee of the First Affiliated Hospital of Anhui Medical University. Each participant signed the informed consent form.
SAMPLE PREPARATION:
Genomic DNA was extracted from venous blood using the Qiagen Genomic Flexi Gene DNA Kit51206 (Qiagen, Germany). DNA templates used for CNV analysis were prepared as follows: DNA quality was assessed by gel electrophoresis to confirm no DNA degradation had occurred and was then diluted to 20 ng/μl and an OD260/280 ratio range of 1.8–2.0 for utilization.
QUANTIFICATION OF COPY NUMBERS:
Copy number was determined using the 2−ΔΔCTmethod using TaqMan copy number targets and RNase P reference assays (assay ID: 4403328). TaqMan Copy Number Assays Hs00146600_cn (for
STATISTICAL ANALYSIS:
Statistical analysis was performed using the R 3.4.0 program for Windows (
Results
CLINICAL DATA OF ALL PARTICIPANTS:
Patients were classified as PsV (n=283), PsA (n=74), EP (n=69), and GPP (n=52). The mean age of psoriasis patients and healthy controls was 39.6±16.1 and 35.7±16.6 years, respectively. The characteristics of subjects in the study are shown in Table 1. There was no significant difference in the distribution of male and female subjects between case and control cohorts (P>0.01).
CNV OF IL22 IN PSORIASIS SUBTYPES:
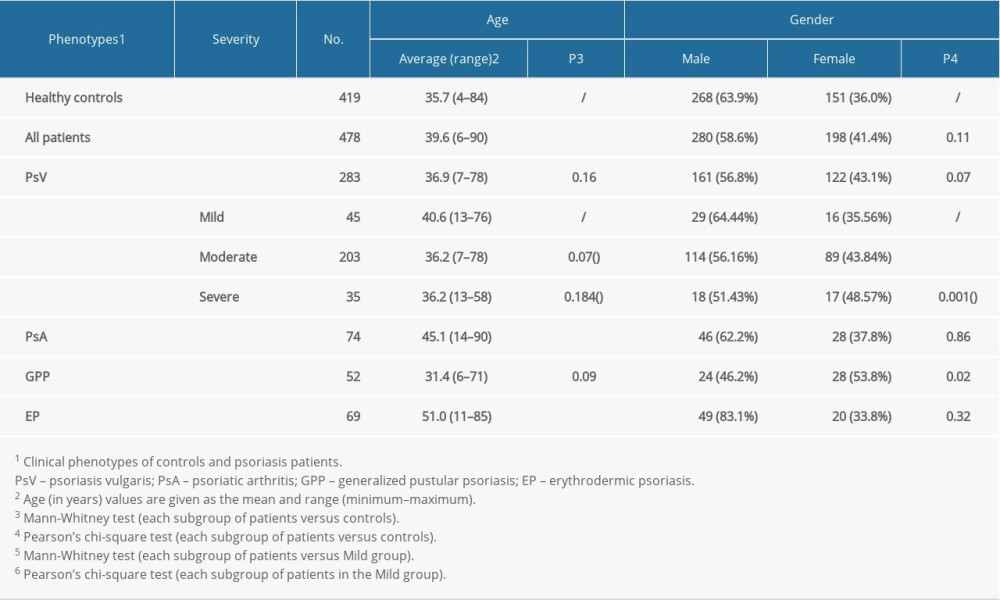
Copy numbers of the IL22 gene ranged from 2 to 7 (mean±SD, 2.31±0.6) in controls and 2 to 5 (mean±SD, 2.36±0.59) in patients. Mean copy numbers of IL22 were significantly higher in the PsV group compared to the control group (P=0.001, t test) (Figure 1A). No strong association was observed in comparisons between PsA, GPP, or EP and controls (Figure 1A). Allele frequency of IL22 copy numbers significantly differed in cases and controls, with 35% patients carried 3 or more copies, while only 26.3% controls obtained more than 3 copies. (P=0.021, Pearson’s χ2 test; OR=0.036, 95% CI=0.18–0.82) (Figure 2A). Furthermore, those who had more than 2 copies of IL22 were more likely to develop PsV (P=0.028, Figure 2B), suggesting IL22 copy numbers increased risk of affecting psoriasis.
We then looked at copy number alterations among psoriasis subtypes. Mean copy numbers of IL22 were higher in PsV than that in PsA groups (P=0.0025), but not in other subtypes (Figure 1A). Interestingly, PsV group presented wider copy number spectrums than PsA group, with 2–6 copies in PsV and 2–3 copies in PsA (Figure 2D). A distinct distribution was observed between the PsV and PsA groups (P=7.75E-10, OR=0.089, 95% CI=0.26–2.83, Figure 2C). These findings revealed IL22 copy numbers might differentially associated with psoriasis subtypes.
CNV OF LCE3C IN PSORIASIS SUBTYPES:
The mean LCE3C copy number in the PsV group was significantly lower when compared to the control group (P<0.001) (Figure 1B). The copy number distribution difference was observed between PsV and control groups (P<0.001, OR=1.4, 95% CI=0.91–2.18, Figure 3A). Furthermore, those who had 2 copies of LCE3C were more likely to develop PsV (P=0.0018, Figure 3B). The mean LCE3C copy number in PsA and GPP groups was lower compared to the control group (P=0.0046 and P=1.56E-04, respectively) (Figure 1B). The same distribution trend was observed when comparing the GPP and control groups (P=0.078, OR=0.13, 95% CI=0.06–0.31) (Figure 3C). Those who had 2 copies of LCE3C were more probably to develop GPP (P<0.05, Figure 3D). The copy numbers of LCE3C tended to be associated with EP, although the comparison did not reach significance (P=0.076).
CNV OF IL22 LINKED WITH PSORIASIS SEVERITY:
Given the critical roles of IL22 in psoriasis development, we evaluated the relationship between CNV of IL22 and clinical parameters of psoriasis. We found patients with different IL22 copy numbers obtained varied severity scores (mild, moderate, and severe) (Figure 4A). We also found the mean BSA scores were dramatically higher in patients with 2 copy numbers than that in 3 or more carriers, and the CNV of IL22 was inversely correlated with the severity of psoriasis (P=0.0348, R2=0.9316,Figure 4A). CNV of IL22 in the mild group were significantly higher than in either the severe group or healthy controls (Mild: 2.53±0.694, Severe: 2.33±0.581, Control: 2.31±0.598, PMild vs Severe=0.043, PMild vs Control=0.019, respectively, Figure 4C). We also evaluated the relationship between CNV in the LCE3C gene and the clinical parameters of psoriasis. Similar to IL22 copy numbers, BSA scores varied in patients with different LCE3C copy numbers (Figure 4B). But the correlation between CNV of LCE3C and the trends of BSA score were not found (Figure 4B). However, we noticed that CNV of LCE3C in mild, moderate and severe psoriasis being lower compared to healthy controls (Pmild=0.007, Pmoderate=0.002, Psevere<0.001, respectively, Figure 4D).
For genetic background would be associated with early onset of psoriasis [32]. We assessed whether CNVs of IL22 and LEC3C were risk factors for early onset, by comparing the copy number allele frequencies in early and late-onset 4 subtypes of psoriasis. No significant relationships were observed, indicating CNVs of IL22 and LEC3C are not the key predisposing factors for early onset of psoriasis.
Discussion
Psoriasis is a common skin disorder with strong environmental and genetic components. Genome variation studies have revealed at least 100 disease-associated risk loci or genes, including CNVs in
In this study, CNVs of
We also identified differential distribution of
The
Conclusions
In conclusion, we observed that CNV of
Further research is needed to determine whether CNVs of
Figures
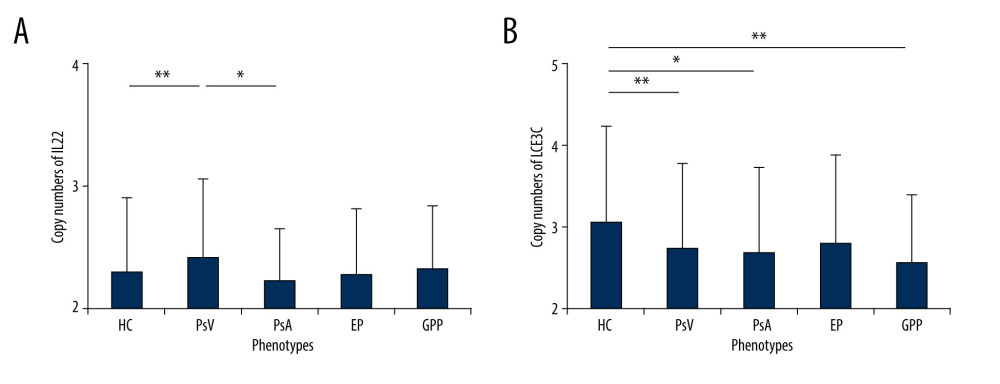 Figure 1. Frequency distributions of IL22 and LCE3C CNVs. (A) Associations of IL22 gene copy number with PsV. (B) LCE3C CNVs are associated with PsV, PsA and GPP. ** P<0.001, * P<0.05. HC – healthy control; PsV – psoriasis vulgaris; PsA – psoriatic arthritis; EP – erythrodermic psoriasis; GPP – generalized pustular psoriasis. (The figure was created by Microsoft Excel 2019 software, Microsoft, USA).
Figure 1. Frequency distributions of IL22 and LCE3C CNVs. (A) Associations of IL22 gene copy number with PsV. (B) LCE3C CNVs are associated with PsV, PsA and GPP. ** P<0.001, * P<0.05. HC – healthy control; PsV – psoriasis vulgaris; PsA – psoriatic arthritis; EP – erythrodermic psoriasis; GPP – generalized pustular psoriasis. (The figure was created by Microsoft Excel 2019 software, Microsoft, USA). 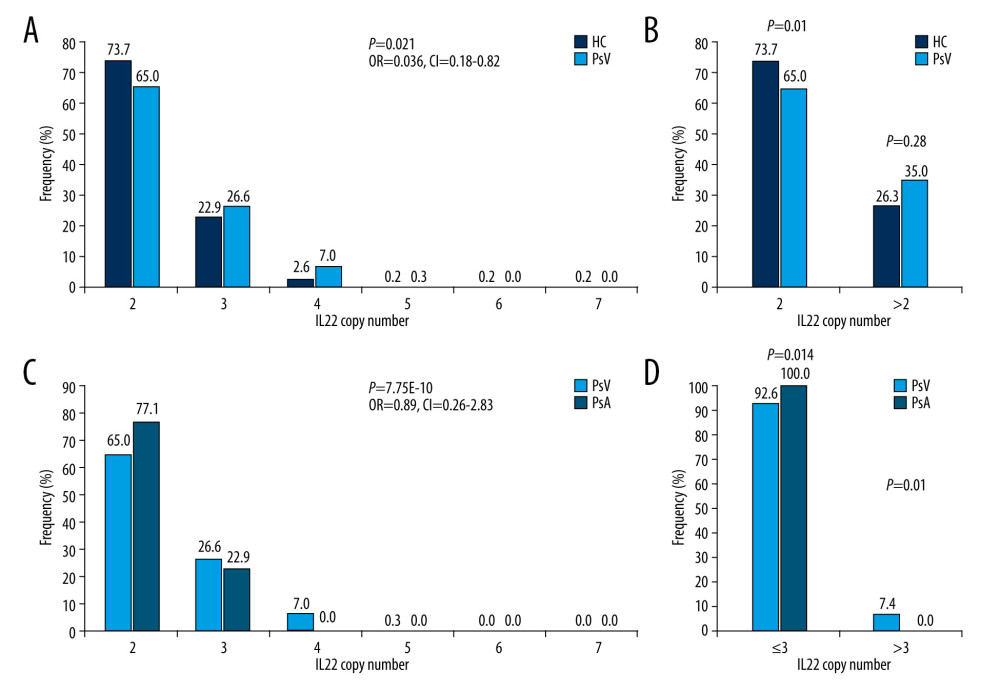 Figure 2. Distribution of IL22 in PsV, PsA and HC. (A) IL22 copy numbers were significantly different between PsV and HC. (B) More than 2 copies of IL22 increased the risk of developing PsV. (C) PsA has lower IL22 copy number compared to PsV. (D) The PsV group had wider IL22 copy number spectrums than the PsA group. HC – healthy control; PsV – psoriasis vulgaris; PsA – psoriatic arthritis. (The figure was created by Microsoft Excel 2019 software, Microsoft, USA).
Figure 2. Distribution of IL22 in PsV, PsA and HC. (A) IL22 copy numbers were significantly different between PsV and HC. (B) More than 2 copies of IL22 increased the risk of developing PsV. (C) PsA has lower IL22 copy number compared to PsV. (D) The PsV group had wider IL22 copy number spectrums than the PsA group. HC – healthy control; PsV – psoriasis vulgaris; PsA – psoriatic arthritis. (The figure was created by Microsoft Excel 2019 software, Microsoft, USA).  Figure 3. Distribution of LCE3C in PsV, GPP, and HC. (A) LCE3C copy numbers were significantly different between PsV and HC. (B) More than 2 copies of LCE3C decrease the risk of developing PsV. (C) LCE3C copy numbers were significantly different between GPP and HC. (D) More than 2 copies of LCE3C decreased the risk of developing GPP. HC – healthy control; PsV – psoriasis vulgaris; GPP – generalized pustular psoriasis. (The figure was created by Microsoft Excel 2019 software, Microsoft, USA).
Figure 3. Distribution of LCE3C in PsV, GPP, and HC. (A) LCE3C copy numbers were significantly different between PsV and HC. (B) More than 2 copies of LCE3C decrease the risk of developing PsV. (C) LCE3C copy numbers were significantly different between GPP and HC. (D) More than 2 copies of LCE3C decreased the risk of developing GPP. HC – healthy control; PsV – psoriasis vulgaris; GPP – generalized pustular psoriasis. (The figure was created by Microsoft Excel 2019 software, Microsoft, USA). 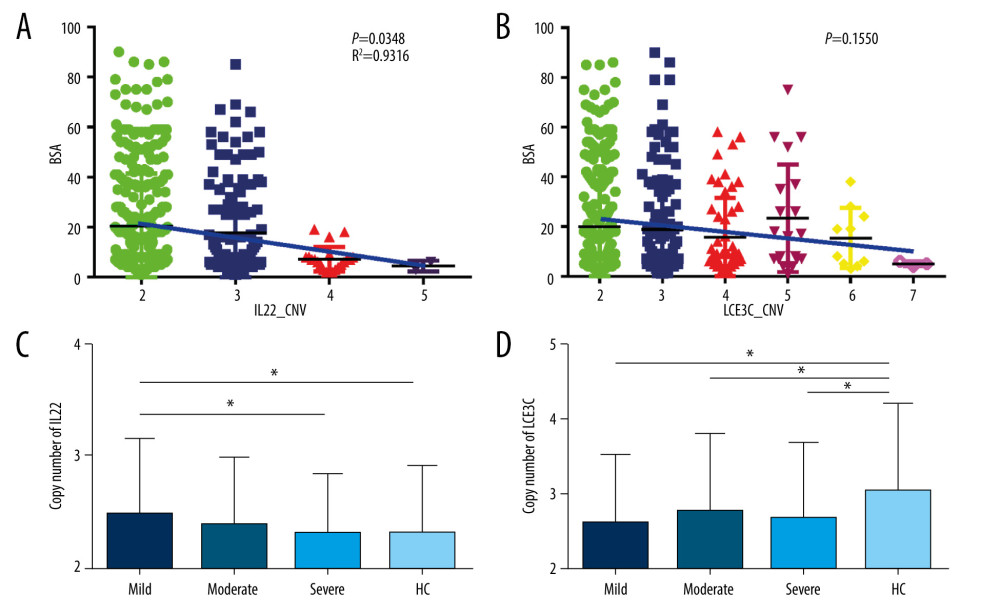 Figure 4. Associations of IL22 and LCE3C copy numbers among different severities of psoriasis and healthy controls. (A) The distribution of IL22 CNV was different and inversely correlated with the severity of psoriasis. (B) The distribution of LCE3C CNV was different and inversely correlated with the severity of psoriasis. (C) IL22 copy numbers in mild psoriasis are higher than in severe psoriasis and in healthy controls. (D) LCE3C copy numbers in mild, moderate, and severe psoriasis were lower than in healthy controls. Mild – mild psoriasis; Moderate – moderate psoriasis; Severe – severe psoriasis; HC – healthy control; Blue Bar – the correlation between different CNV group and the BSA score. * P<0.05. (The figure was created by Microsoft Excel 2019 software, Microsoft, USA).
Figure 4. Associations of IL22 and LCE3C copy numbers among different severities of psoriasis and healthy controls. (A) The distribution of IL22 CNV was different and inversely correlated with the severity of psoriasis. (B) The distribution of LCE3C CNV was different and inversely correlated with the severity of psoriasis. (C) IL22 copy numbers in mild psoriasis are higher than in severe psoriasis and in healthy controls. (D) LCE3C copy numbers in mild, moderate, and severe psoriasis were lower than in healthy controls. Mild – mild psoriasis; Moderate – moderate psoriasis; Severe – severe psoriasis; HC – healthy control; Blue Bar – the correlation between different CNV group and the BSA score. * P<0.05. (The figure was created by Microsoft Excel 2019 software, Microsoft, USA). References
1. Harden JL, Krueger JG, Bowcock AM, The immunogenetics of psoriasis: A comprehensive review: J Autoimmun, 2015; 64; 66-73
2. Nikamo P, Cheuk S, Lysell J, Genetic variants of the IL22 promoter associate to onset of psoriasis before puberty and increased IL-22 production in T cells: J Invest Dermatol, 2014; 134; 1535-41
3. Boehncke WH, Schon MP, Psoriasis: Lancet, 2015; 386; 983-94
4. Palfreeman AC, McNamee KE, McCann FE, New developments in the management of psoriasis and psoriatic arthritis: A focus on apremilast: Drug Des Devel Ther, 2013; 7; 201-10
5. Griffiths CE, Barker JN, Pathogenesis and clinical features of psoriasis: Lancet, 2007; 370; 263-71
6. Griffiths C, Barker J, Bleiker TO: Rook’s textbook of dermatology, 4 Volume Set, 2016, Wiley
7. Moll JM, Wright V, Psoriatic arthritis: Semin Arthritis Rheum, 1973; 3; 55-78
8. Rosenbach M, Hsu S, Korman NJ, Treatment of erythrodermic psoriasis: From the medical board of the National Psoriasis Foundation: J Am Acad Dermatol, 2010; 62; 655-62
9. Fujita H, Terui T, Hayama K, Japanese guidelines for the management and treatment of generalized pustular psoriasis: The new pathogenesis and treatment of GPP: J Dermatol, 2018; 45; 1235-70
10. Navarini AA, Burden AD, Capon F, European consensus statement on phenotypes of pustular psoriasis: J Eur Acad Dermatol Venereol, 2017; 31; 1792-99
11. Hwang ST, Nijsten T, Elder JT, Recent highlights in psoriasis research: J Invest Dermatol, 2017; 137; 550-56
12. Zhang XJ, Huang W, Yang S, Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21: Nat Genet, 2009; 41; 205-10
13. Yin X, Wineinger NE, Cheng H, Common variants explain a large fraction of the variability in the liability to psoriasis in a Han Chinese population: BMC Genomics, 2014; 15; 87
14. Redon R, Ishikawa S, Fitch KR, Global variation in copy number in the human genome: Nature, 2006; 444; 444-54
15. Kleinjan DA, van Heyningen V, Long-range control of gene expression: Emerging mechanisms and disruption in disease: Am J Hum Genet, 2005; 76; 8-32
16. McCarroll SA, Hadnott TN, Perry GH, Common deletion polymorphisms in the human genome: Nat Genet, 2006; 38; 86-92
17. Rodriguez-Revenga L, Mila M, Rosenberg C, Structural variation in the human genome: The impact of copy number variants on clinical diagnosis: Genet Med, 2007; 9; 600-6
18. Stranger BE, Forrest MS, Dunning M, Relative impact of nucleotide and copy number variation on gene expression phenotypes: Science, 2007; 315; 848-53
19. Wu Y, Zhang Z, Tao L, A high copy number of FCGR3B is associated with psoriasis vulgaris in Han Chinese: Dermatology, 2014; 229; 70-75
20. Hollox EJ, Huffmeier U, Zeeuwen PL, Psoriasis is associated with increased beta-defensin genomic copy number: Nat Genet, 2008; 40; 23-25
21. Bassaganyas L, Riveira-Munoz E, Garcia-Aragones M, Worldwide population distribution of the common LCE3C-LCE3B deletion associated with psoriasis and other autoimmune disorders: BMC Genomics, 2013; 14; 261
22. de Cid R, Riveira-Munoz E, Zeeuwen PL, Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis: Nat Genet, 2009; 41; 211-15
23. Li M, Wu Y, Chen G, Deletion of the late cornified envelope genes LCE3C and LCE3B is associated with psoriasis in a Chinese population: J Invest Dermatol, 2011; 131; 1639-43
24. Prans E, Kingo K, Traks T, Copy number variations in IL22 gene are associated with Psoriasis vulgaris: Hum Immunol, 2013; 74; 792-95
25. Zhou F, Shen C, Hsu YH, DNA methylation-based subclassification of psoriasis in the Chinese Han population: Front Med, 2018; 12; 717-25
26. Bashir S, Hassan I, Majid S, Feasibility of establishing deletion of the late cornified envelope genes LCE3B and LCE3C as a susceptibility factor for psoriasis: Adv Biomed Res, 2016; 5; 109
27. Coin LJ, Cao D, Ren J, An exome sequencing pipeline for identifying and genotyping common CNVs associated with disease with application to psoriasis: Bioinformatics, 2012; 28; i370-i74
28. Huffmeier U, Bergboer JG, Becker T, Replication of LCE3C-LCE3B CNV as a risk factor for psoriasis and analysis of interaction with other genetic risk factors: J Invest Dermatol, 2010; 130; 979-84
29. Taylor W, Gladman D, Helliwell P, Classification criteria for psoriatic arthritis: Development of new criteria from a large international study: Arthritis Rheum, 2006; 54; 2665-73
30. Menter A, Strober BE, Kaplan DH, Joint AAD-NPF guidelines of care for the management and treatment of psoriasis with biologics: J Am Acad Dermatol, 2019; 80; 1029-72
31. Knuckles MLF, Levi E, Soung J, Defining and treating moderate plaque psoriasis: A dermatologist survey: J Dermatolog Treat, 2018; 29; 658-63
32. Sun LD, Cheng H, Wang ZX, Association analyses identify six new psoriasis susceptibility loci in the Chinese population: Nat Genet, 2010; 42; 1005-9
33. Sonnenberg GF, Fouser LA, Artis D, Border patrol: Regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22: Nat Immunol, 2011; 12; 383-90
34. Ma HL, Liang S, Li J, IL-22 is required for Th17 cell-mediated pathology in a mouse model of psoriasis-like skin inflammation: J Clin Invest, 2008; 118; 597-607
35. Mitra A, Raychaudhuri SK, Raychaudhuri SP, Functional role of IL-22 in psoriatic arthritis: Arthritis Res Ther, 2012; 14; R65
36. Zheng Y, Danilenko DM, Valdez P, Interleukin-22, a T(H)17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis: Nature, 2007; 445; 648-51
37. Wang B, Han D, Li F, Elevated IL-22 in psoriasis plays an anti-apoptotic role in keratinocytes through mediating Bcl-xL/Bax: Apoptosis, 2020; 25; 663-73
38. Yamamoto M, Imai Y, Sakaguchi Y, Serum cytokines correlated with the disease severity of generalized pustular psoriasis: Dis Markers, 2013; 34; 153-61
39. Yu B, Guan M, Peng YH, Copy number variations of interleukin-17F, interleukin-21, and interleukin-22 are associated with systemic lupus erythematosus: Arthritis Rheum, 2011; 63; 3487-92
40. Tokuyama M, Mabuchi T, New treatment addressing the pathogenesis of psoriasis: Int J Mol Sci, 2020; 21; 7488
41. Sbidian E, Chaimani A, Afach S, Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis: Cochrane Database Syst Rev, 2020; 1; CD011535
42. Bao L, Li J, Perez White BE, Inhibition of dipeptidyl-peptidase 4 induces upregulation of the late cornified envelope cluster in keratinocytes: Arch Dermatol Res, 2021 [Online ahead of print]
43. Riveira-Munoz E, He SM, Escaramis G, Meta-analysis confirms the LCE3C_LCE3B deletion as a risk factor for psoriasis in several ethnic groups and finds interaction with HLA-Cw6: J Invest Dermatol, 2011; 131; 1105-9
Figures
 Figure 1. Frequency distributions of IL22 and LCE3C CNVs. (A) Associations of IL22 gene copy number with PsV. (B) LCE3C CNVs are associated with PsV, PsA and GPP. ** P<0.001, * P<0.05. HC – healthy control; PsV – psoriasis vulgaris; PsA – psoriatic arthritis; EP – erythrodermic psoriasis; GPP – generalized pustular psoriasis. (The figure was created by Microsoft Excel 2019 software, Microsoft, USA).
Figure 1. Frequency distributions of IL22 and LCE3C CNVs. (A) Associations of IL22 gene copy number with PsV. (B) LCE3C CNVs are associated with PsV, PsA and GPP. ** P<0.001, * P<0.05. HC – healthy control; PsV – psoriasis vulgaris; PsA – psoriatic arthritis; EP – erythrodermic psoriasis; GPP – generalized pustular psoriasis. (The figure was created by Microsoft Excel 2019 software, Microsoft, USA). Figure 2. Distribution of IL22 in PsV, PsA and HC. (A) IL22 copy numbers were significantly different between PsV and HC. (B) More than 2 copies of IL22 increased the risk of developing PsV. (C) PsA has lower IL22 copy number compared to PsV. (D) The PsV group had wider IL22 copy number spectrums than the PsA group. HC – healthy control; PsV – psoriasis vulgaris; PsA – psoriatic arthritis. (The figure was created by Microsoft Excel 2019 software, Microsoft, USA).
Figure 2. Distribution of IL22 in PsV, PsA and HC. (A) IL22 copy numbers were significantly different between PsV and HC. (B) More than 2 copies of IL22 increased the risk of developing PsV. (C) PsA has lower IL22 copy number compared to PsV. (D) The PsV group had wider IL22 copy number spectrums than the PsA group. HC – healthy control; PsV – psoriasis vulgaris; PsA – psoriatic arthritis. (The figure was created by Microsoft Excel 2019 software, Microsoft, USA). Figure 3. Distribution of LCE3C in PsV, GPP, and HC. (A) LCE3C copy numbers were significantly different between PsV and HC. (B) More than 2 copies of LCE3C decrease the risk of developing PsV. (C) LCE3C copy numbers were significantly different between GPP and HC. (D) More than 2 copies of LCE3C decreased the risk of developing GPP. HC – healthy control; PsV – psoriasis vulgaris; GPP – generalized pustular psoriasis. (The figure was created by Microsoft Excel 2019 software, Microsoft, USA).
Figure 3. Distribution of LCE3C in PsV, GPP, and HC. (A) LCE3C copy numbers were significantly different between PsV and HC. (B) More than 2 copies of LCE3C decrease the risk of developing PsV. (C) LCE3C copy numbers were significantly different between GPP and HC. (D) More than 2 copies of LCE3C decreased the risk of developing GPP. HC – healthy control; PsV – psoriasis vulgaris; GPP – generalized pustular psoriasis. (The figure was created by Microsoft Excel 2019 software, Microsoft, USA). Figure 4. Associations of IL22 and LCE3C copy numbers among different severities of psoriasis and healthy controls. (A) The distribution of IL22 CNV was different and inversely correlated with the severity of psoriasis. (B) The distribution of LCE3C CNV was different and inversely correlated with the severity of psoriasis. (C) IL22 copy numbers in mild psoriasis are higher than in severe psoriasis and in healthy controls. (D) LCE3C copy numbers in mild, moderate, and severe psoriasis were lower than in healthy controls. Mild – mild psoriasis; Moderate – moderate psoriasis; Severe – severe psoriasis; HC – healthy control; Blue Bar – the correlation between different CNV group and the BSA score. * P<0.05. (The figure was created by Microsoft Excel 2019 software, Microsoft, USA).
Figure 4. Associations of IL22 and LCE3C copy numbers among different severities of psoriasis and healthy controls. (A) The distribution of IL22 CNV was different and inversely correlated with the severity of psoriasis. (B) The distribution of LCE3C CNV was different and inversely correlated with the severity of psoriasis. (C) IL22 copy numbers in mild psoriasis are higher than in severe psoriasis and in healthy controls. (D) LCE3C copy numbers in mild, moderate, and severe psoriasis were lower than in healthy controls. Mild – mild psoriasis; Moderate – moderate psoriasis; Severe – severe psoriasis; HC – healthy control; Blue Bar – the correlation between different CNV group and the BSA score. * P<0.05. (The figure was created by Microsoft Excel 2019 software, Microsoft, USA). In Press
05 Mar 2024 : Clinical Research
Role of Critical Shoulder Angle in Degenerative Type Rotator Cuff Tears: A Turkish Cohort StudyMed Sci Monit In Press; DOI: 10.12659/MSM.943703
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
21 Mar 2024 : Meta-Analysis
Economic Evaluation of COVID-19 Screening Tests and Surveillance Strategies in Low-Income, Middle-Income, a...Med Sci Monit In Press; DOI: 10.12659/MSM.943863
10 Apr 2024 : Clinical Research
Predicting Acute Cardiovascular Complications in COVID-19: Insights from a Specialized Cardiac Referral Dep...Med Sci Monit In Press; DOI: 10.12659/MSM.942612
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952